
Volunteer Distributed Grid Network Computing
Data Science / Distributed Computing / Medical Research
Case Study: Distributed Grid Network Computing for Accelerated Drug Discovery in COVID-19
Client: BALANCED Media | Technology
Objective Amidst the COVID-19 pandemic, finding treatments swiftly was critical to mitigating its global health impact. BALANCED Media | Technology aimed to leverage distributed grid computing through their HEWMEN Cell app, crowd-sourcing computational power to accelerate drug discovery by identifying molecular compounds that could potentially treat COVID-19.
Challenge In late 2019, the novel SARS-CoV-2 virus triggered an unprecedented global crisis. The traditional drug discovery process, often time-consuming, was insufficient to meet the urgent demand for treatments. To address this, BALANCED Media | Technology sought to employ cutting-edge distributed computing techniques, crowd-sourcing computational power from users worldwide to quickly analyze potential drug candidates.
Solution BALANCED Media | Technology, in collaboration with Southern Methodist University’s (SMU) Intelligent Data Analysis Lab, released a specialized version of their HEWMEN Cell app. This app utilized a Distributed Grid Network Computing model, allowing users to volunteer their devices’ computational resources to run simulations aimed at identifying viable treatments for COVID-19.
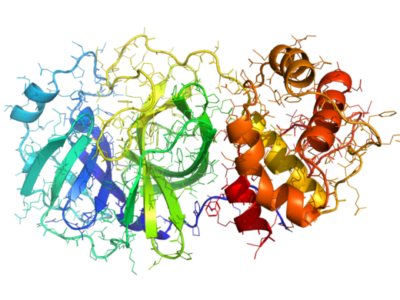
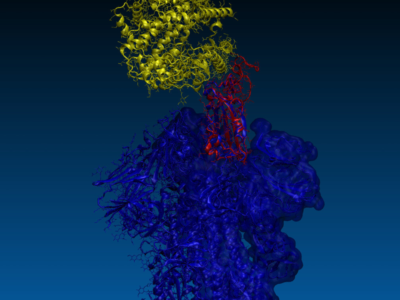
Each user’s device contributed to running molecular docking experiments, simulating how different compounds interacted with viral proteins. The focus was on the SARS-CoV-2 spike protein and the Mpro protein, a crucial target for inhibiting viral replication. The large-scale participation enabled researchers to test vast numbers of compounds faster than traditional methods allowed.
Results HEWMEN Cell’s distributed network processed over 50,000 computational tasks within a few months, surpassing milestones and expediting drug candidate identification. This led to the discovery of promising FDA-approved compounds that could potentially be repurposed for treating COVID-19. These findings were a direct result of the community-driven computational efforts.
Notably, the project identified two targetable sites on the Mpro protein, enabling further research into inhibitors that could disrupt the virus’s ability to replicate. The success of this initiative highlighted how distributed computing could compress years of drug discovery into months, proving vital during a global health emergency.
Impact The project not only advanced COVID-19 treatment research but also demonstrated the potential of distributed grid computing in biomedical research. Thousands of volunteers participated in the initiative, showcasing the power of collective action in solving complex scientific challenges. The project was wound down in February 2022 after successfully achieving its research objectives, but BALANCED Media | Technology continues to explore new applications of this technology in other domains.
By decentralizing computational power and involving the community, BALANCED Media | Technology provided a scalable, efficient model for addressing future public health crises and large-scale scientific problems.
Key Takeaways
- Over 50,000 tasks processed, accelerating drug discovery efforts during COVID-19.
- Identified key target proteins and FDA-approved compounds for further study.
- Leveraged distributed grid computing to crowd-source computational resources.
- Demonstrated the potential of community-powered science in accelerating discovery.


